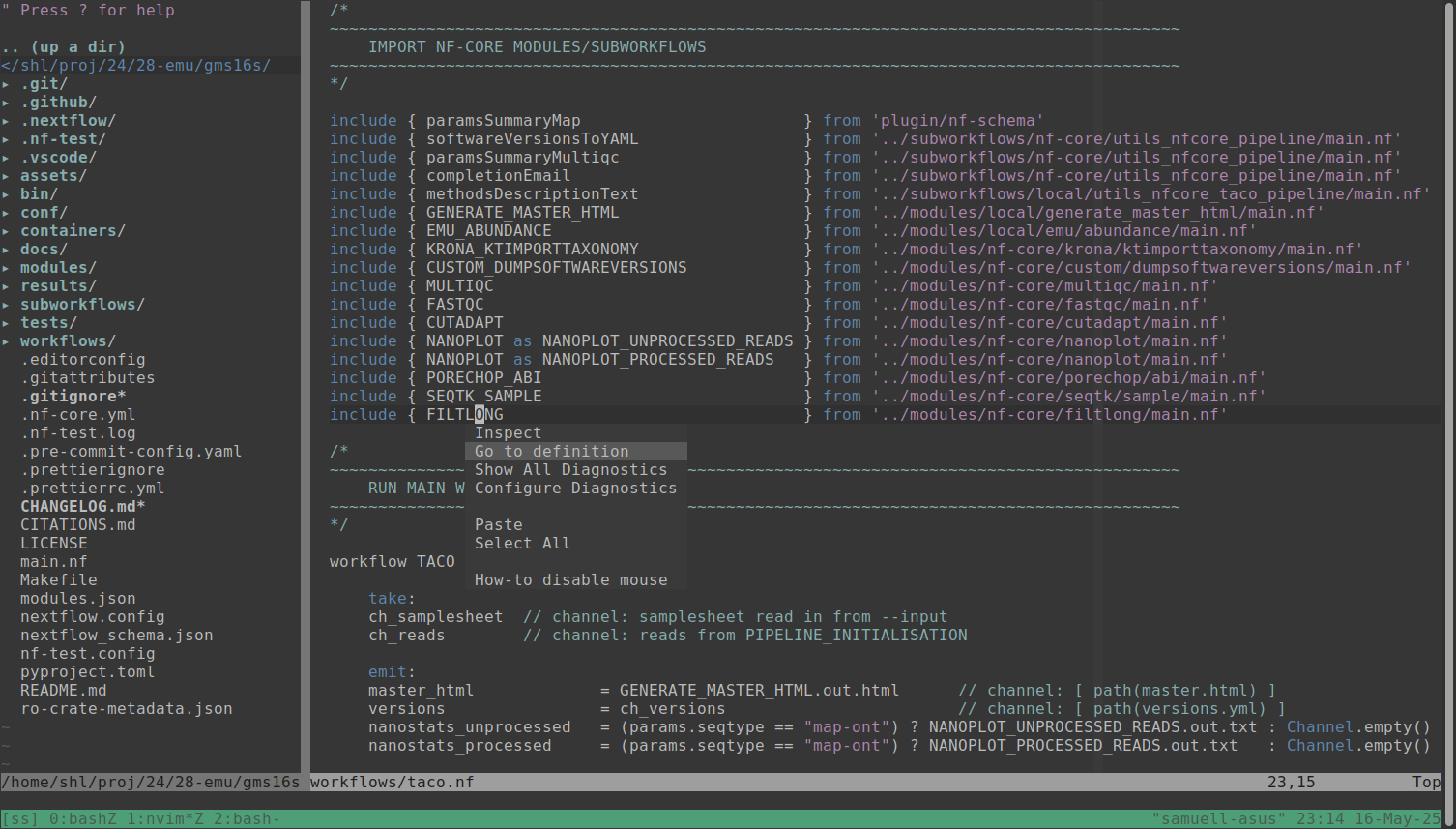
It turns out it is super easy to get Nextflow code intelligence to work in NeoVim now! This is thanks to the newly released Nextflow language server following the increasingly popular Language Server Protocol (LSP).
See this episode of the Nextflow podcast for a more in-depth discussion on this new functionality.
The …
We have been evaluating Nextflow before in my work at pharmb.io , but that was before DSL2 and the support for re-usable modules (which was one reason we needed to develop our own tools to support our challenges, as explained in the paper ). Thus, there’s definitely some stuff to get into.
Based on my years in …
Update (May 2019): A paper incorporating the below considerations is published:
Björn A Grüning, Samuel Lampa, Marc Vaudel, Daniel Blankenberg, “Software engineering for scientific big data analysis ” GigaScience, Volume 8, Issue 5, May 2019, giz054, https://doi.org/10.1093/gigascience/giz054 There are a …
Workflows and DAGs - Confusion about the concepts Jörgen Brandt tweeted a comment that got me thinking again on something I’ve pondered a lot lately:
“A workflow is a DAG.” is really a weak definition. That’s like saying “A love letter is a sequence of characters.” representation ≠ …
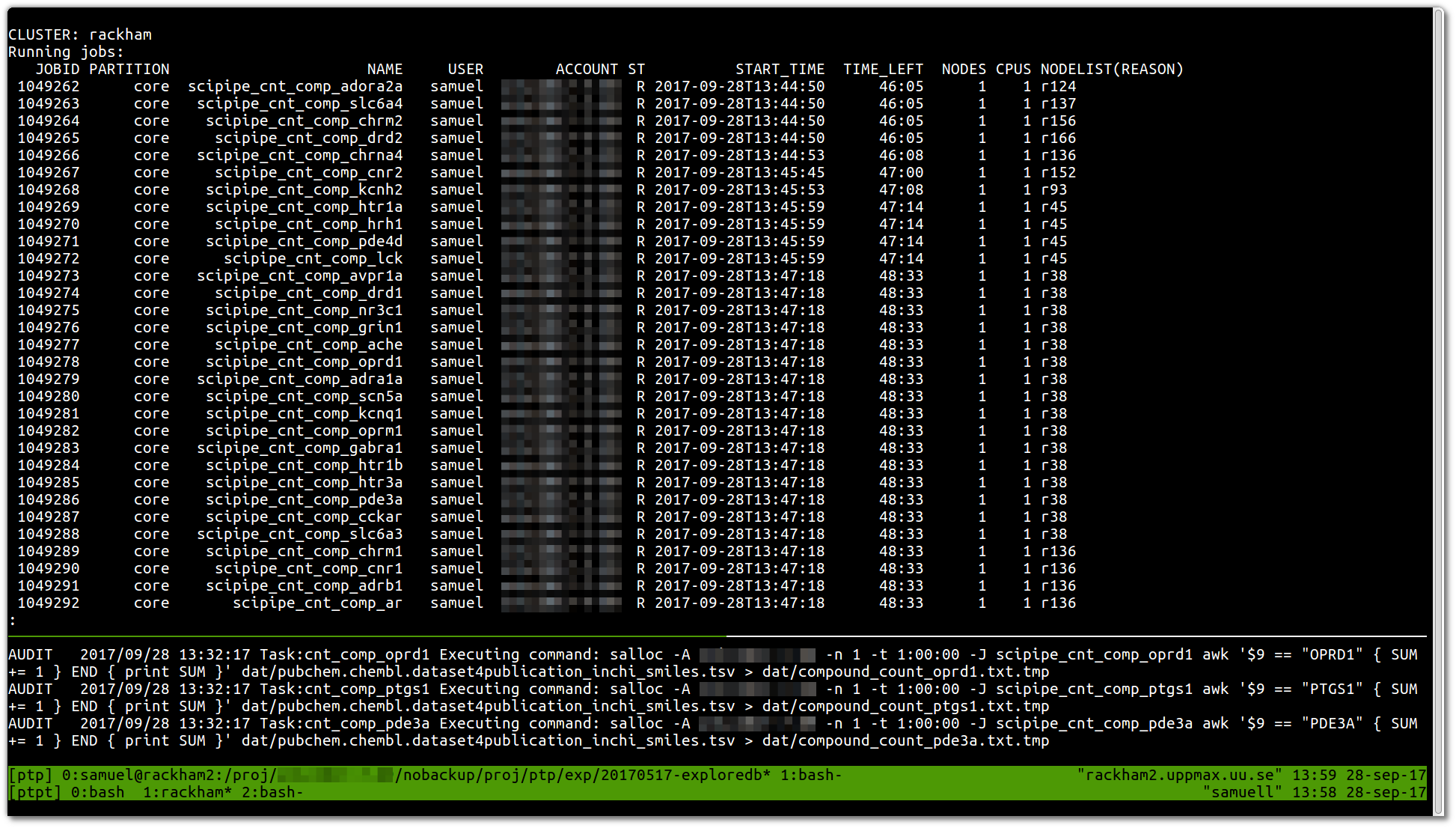
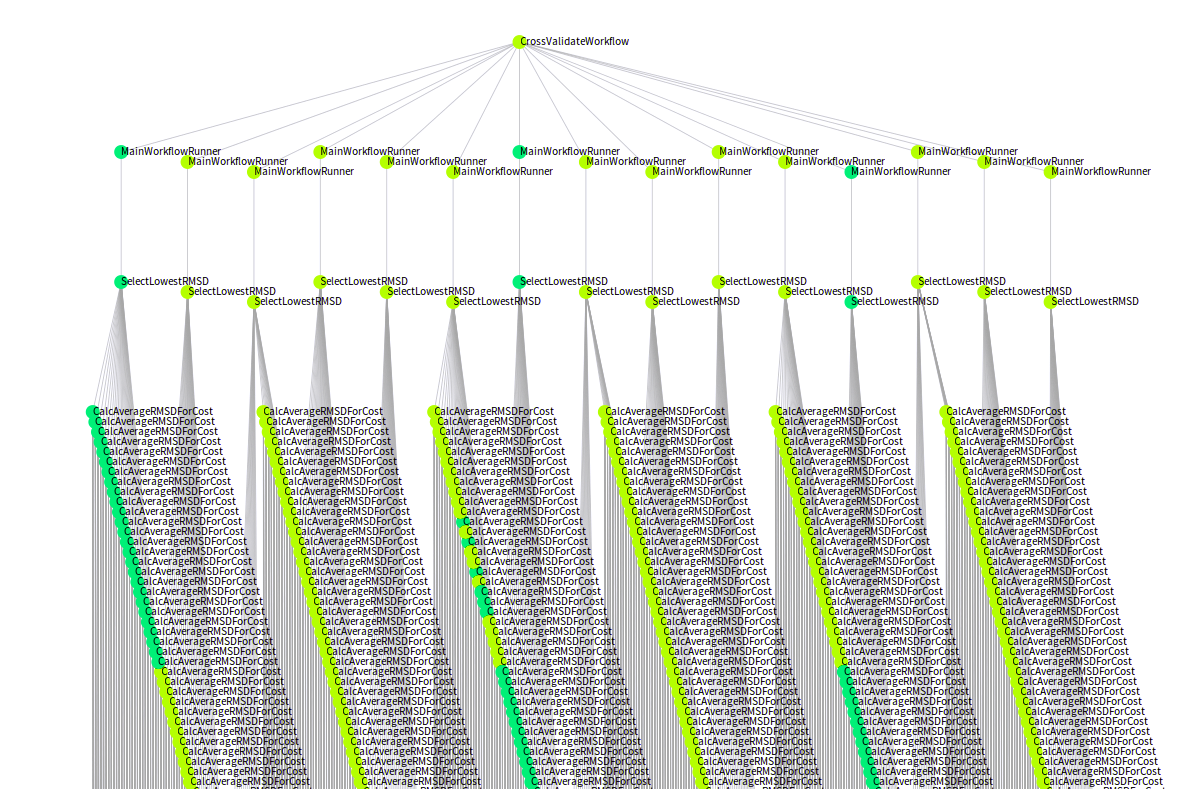
Today marked the day when we ran the very first production workflow with SciPipe , the Go -based scientific workflow tool we’ve been working on over the last couple of years. Yay! :)
This is how it looked (no fancy GUI or such yet, sorry):
The first result we got in this very very first job was a list of counts …
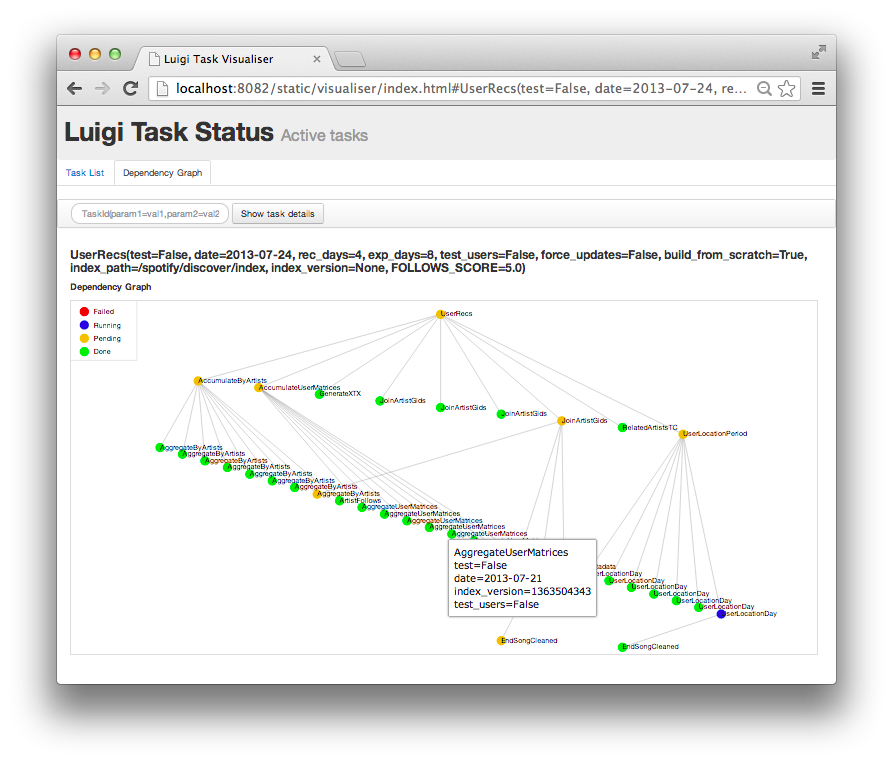
This is a Luigi tutorial I held at the e-Infrastructures for Massively parallel sequencing workshop (Video archive ) at SciLifeLab Uppsala in January 2015, moved here for future reference.
What is Luigi? Luigi is a batch workflow system written in Python and developed by Erik Bernhardson and others at Spotify , where …

Photo credits: Matthew Smith / Unsplash In our work on automating machine learning computations in cheminformatics with scientific workflow tools , we have came to realize something; Dynamic scheduling in scientific workflow tools is very important and sometimes badly needed.
What I mean is that new tasks should be …
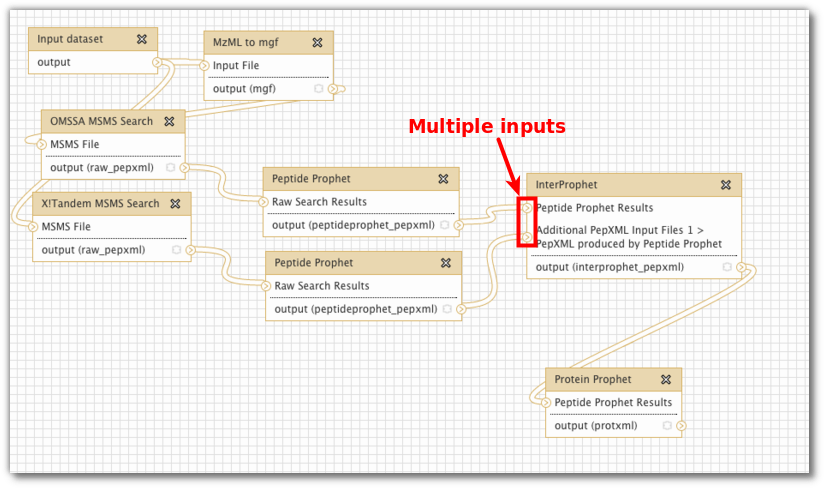
Upsurge in workflow tools There seem to be a little upsurge in light-weight - often python-based - workflow tools for data pipelines in the last couple of years: Spotify’s Luigi , OpenStack’s Mistral , Pinterest’s Pinball , and recently AirBnb’s Airflow , to name a few. These are all interesting …
The workflow problem solved once and for all in 1979? As soon as the topic of scientific workflows is brought up, there are always a few make fans fervently insisting that the problem of workflows is solved once and for all with GNU make , written first in the 70’s :)
Personally I haven’t been so sure. On …
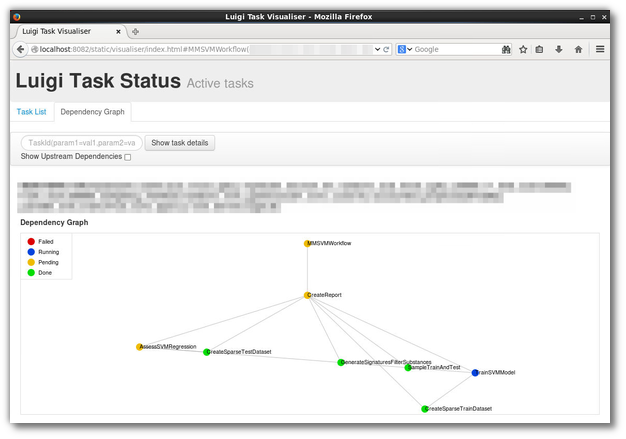
Fig 1: A screenshot of Luigi’s web UI, of a real-world (although rather simple) workflow implemented in Luigi:
Update May 5, 2016: Most of the below material is more or less outdated. Our latest work has resulted in the SciLuigi helper library , which we have used in production and will be focus of further …







