
I was happy to see the publication finally going online , of work done at NASA Glenn Research Center , where SciPipe has been used to process and track provenance of the analyses, “Modeling the impact of thoracic pressure on intracranial pressure”. I’ve known the work existed for a couple of years, …

We just wanted to share that the paper on our Go-based workflow library, SciPipe, was just published in GigaScience:
Abstract Background The complex nature of biological data has driven the development of specialized software tools. Scientific workflow management systems simplify the assembly of such tools into …

A pre-print for our Go-based workflow libarary SciPipe , is out, with the title SciPipe - A workflow library for agile development of complex and dynamic bioinformatics pipelines , co-authored by me and colleagues at pharmb.io : Martin Dahlö , Jonathan Alvarsson and Ola Spjuth . Access it here .
It has been more than …
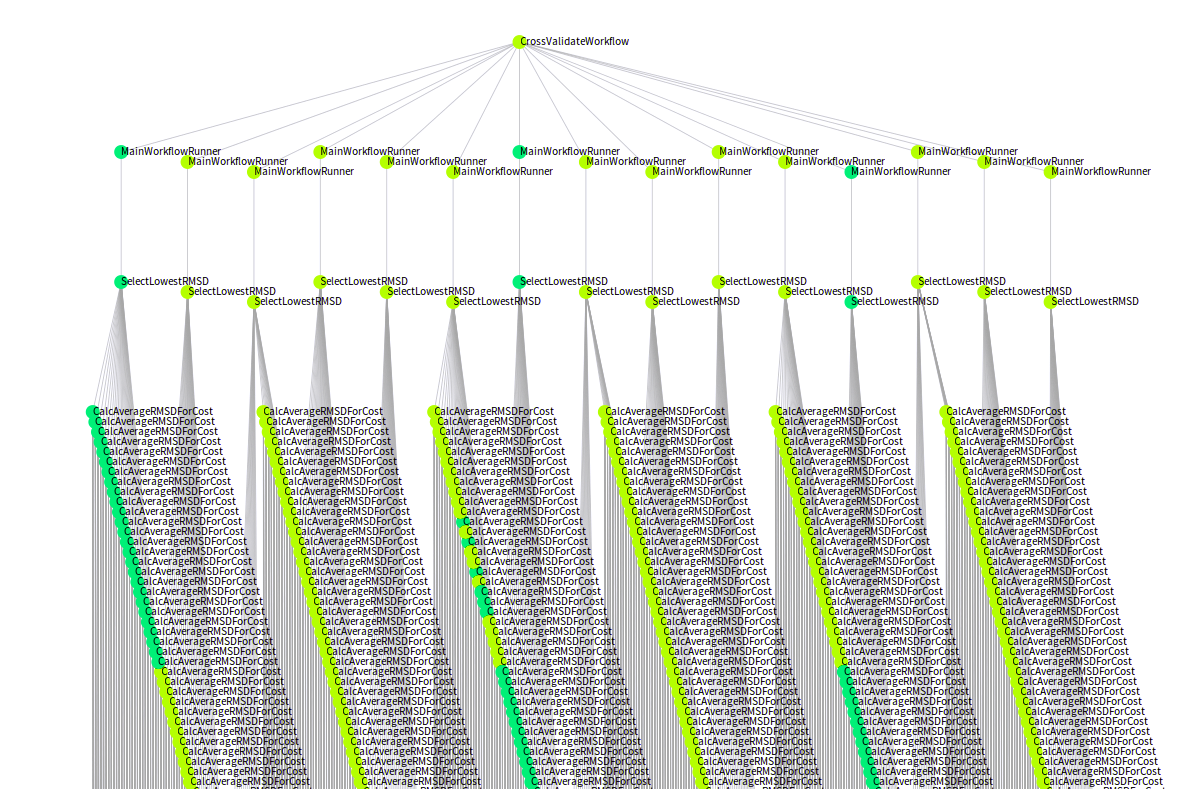
Workflows and DAGs - Confusion about the concepts Jörgen Brandt tweeted a comment that got me thinking again on something I’ve pondered a lot lately:
“A workflow is a DAG.” is really a weak definition. That’s like saying “A love letter is a sequence of characters.” representation ≠ …
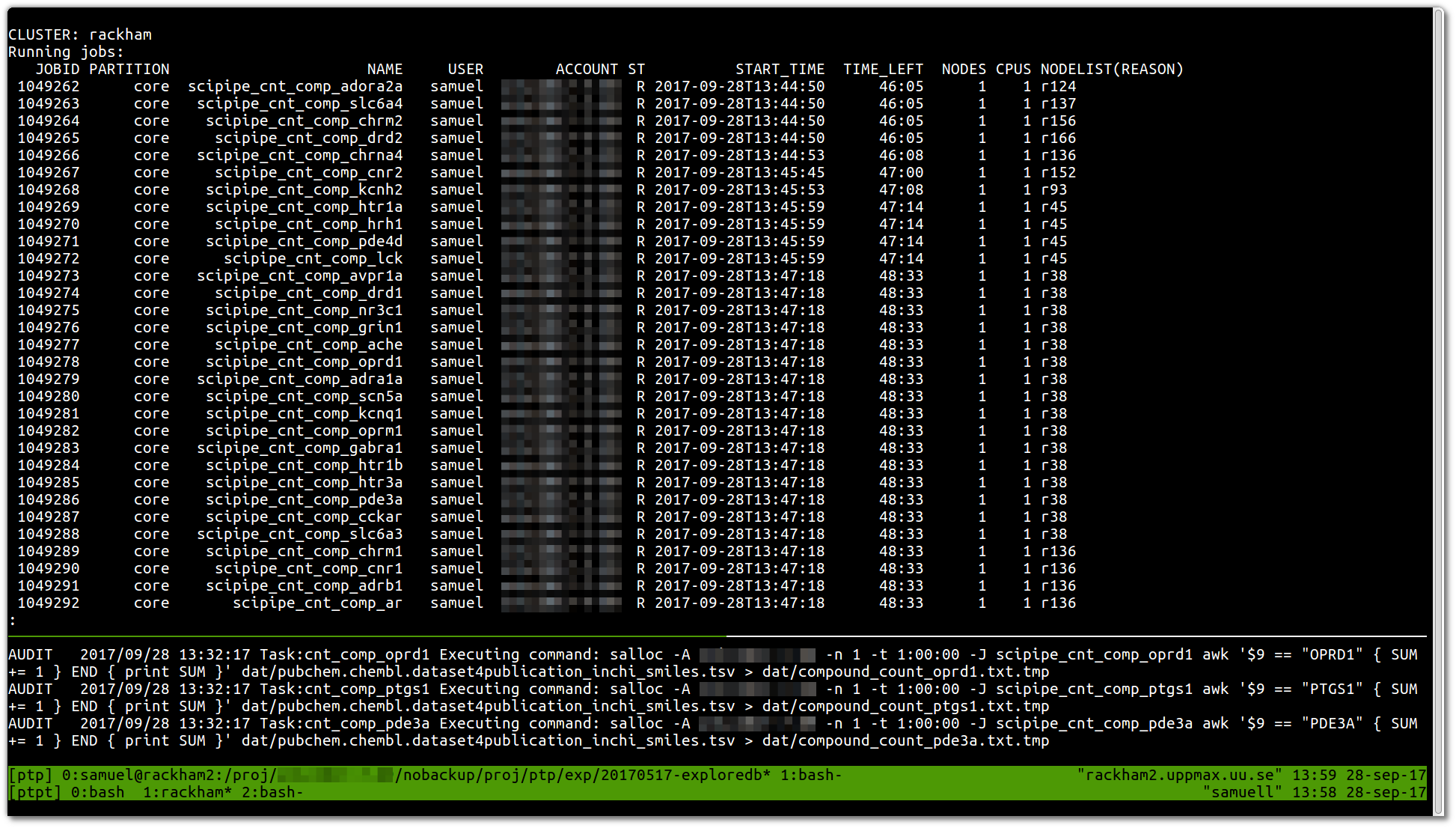
Today marked the day when we ran the very first production workflow with SciPipe , the Go -based scientific workflow tool we’ve been working on over the last couple of years. Yay! :)
This is how it looked (no fancy GUI or such yet, sorry):
The first result we got in this very very first job was a list of counts …




