When I presented a poster about SciCommander at the Swedish bioinformatics workshop last year, I got a lot of awesome feedback from some great people including Fredrik Boulund, Johannes Alneberg and others, of which I unfortunately lost the names (please shout out if you read this!).
(For those new to SciCommander, it …
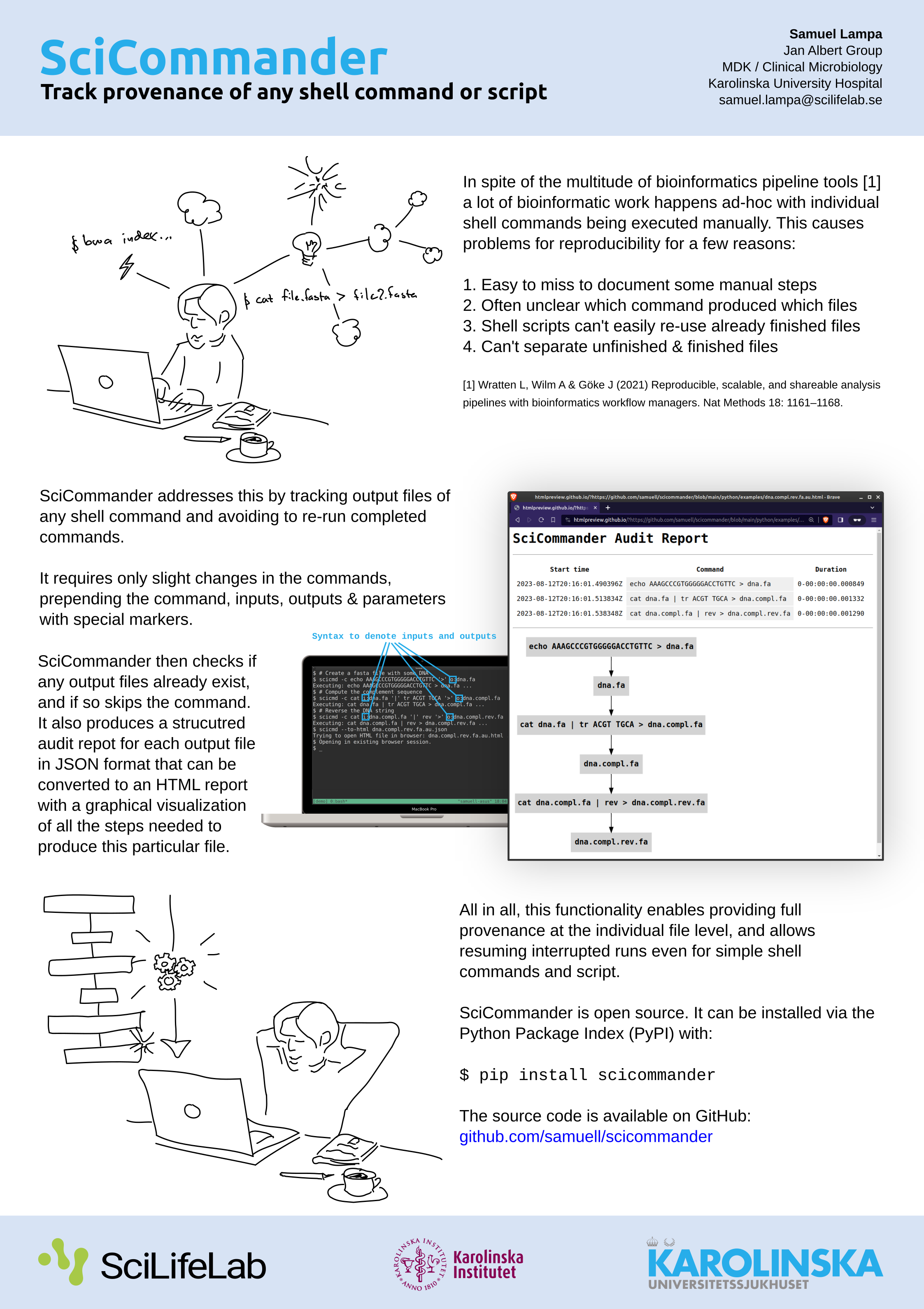
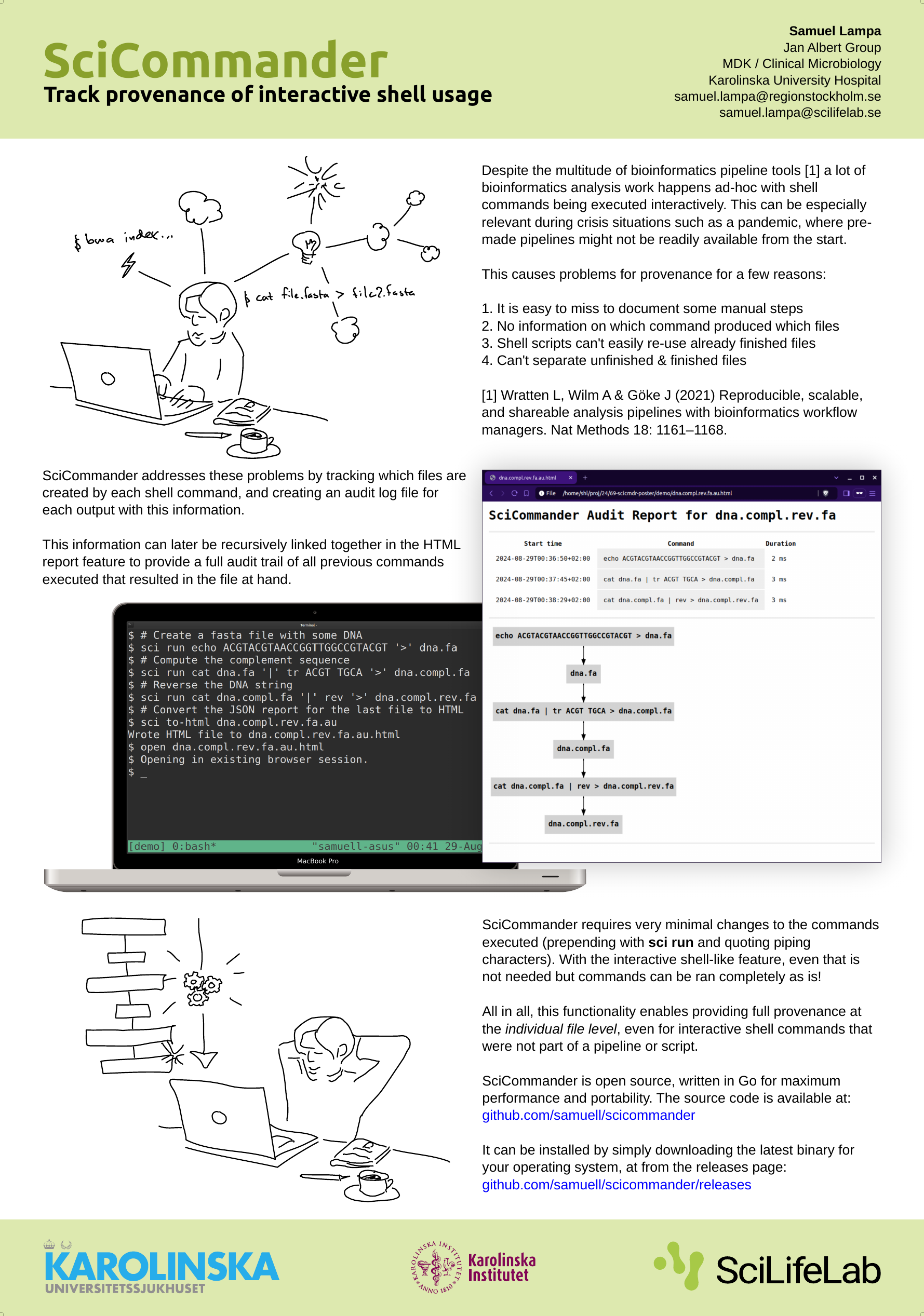
I haven’t written much about a new tool I’ve been working on in some extra time: SciCommander .
I just presented a poster about it at the Swedish Bioinformatics Workshop 2023 , so perhaps let me first present you the poster instead of re-iterating what it is (click to view large version):
New version not …
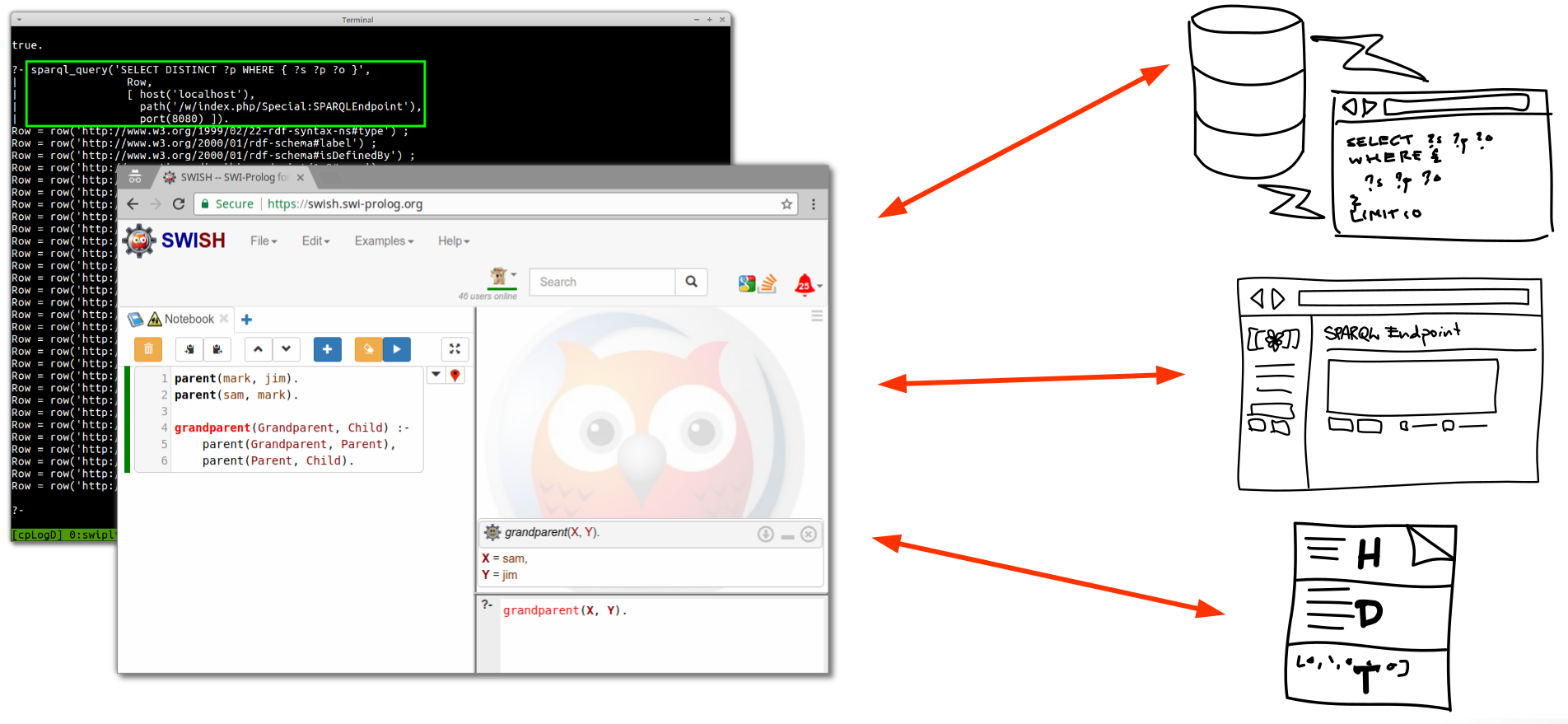
This is an excerpt from the “future outlook” section of my thesis titled “Reproducible Data Analysis in Drug Discovery with Scientific Workflows and the Semantic Web” (click for the open access full text), which aims to provide various putative ways towards improved reproducibility, …
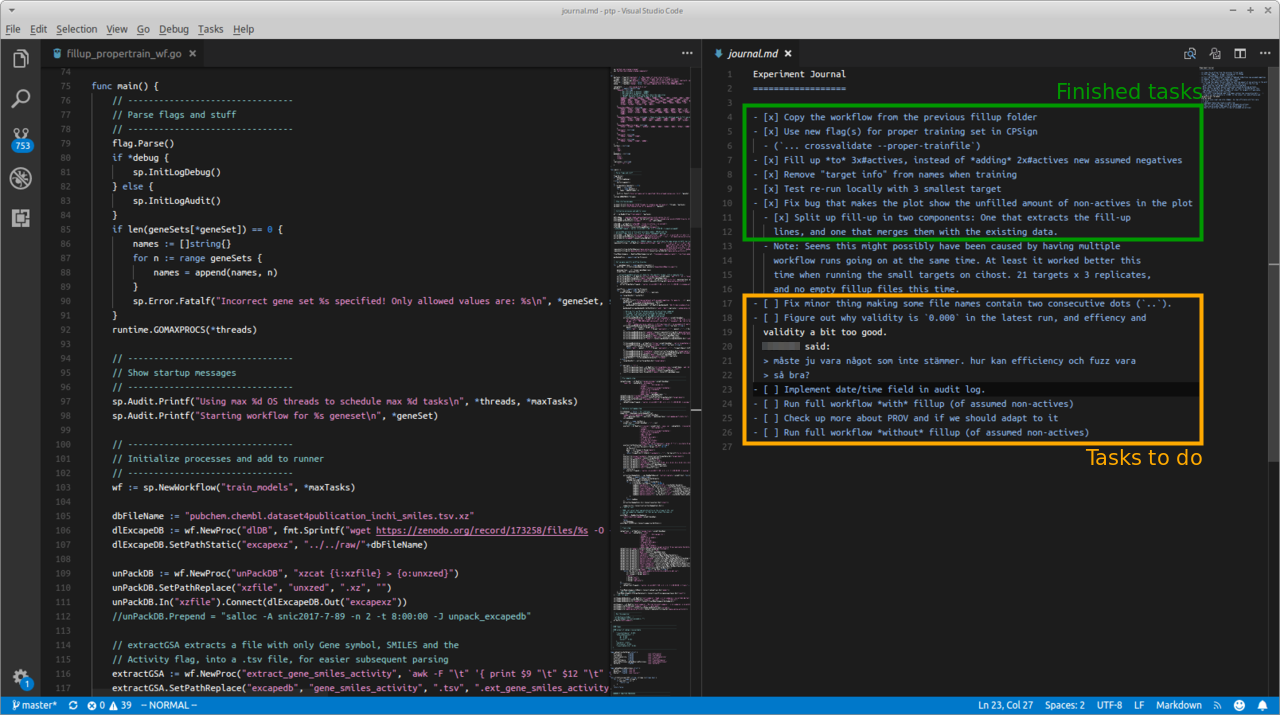
Good lab note-taking is hard Good note-taking is in my opinion as important for computational research as for wet lab research. For computational research it is much easier though to forget doing it, since you might not have a physical notebook lying on your desk staring at you, but rather might need to open a specific …
One of the more important tasks for a scientific workflow is to keep track of so called “provenance information” about its data outputs - information about how each data file was created. This is important so other researchers can easily replicate the study (re-run it with the same software and tools). It …
The workflow problem solved once and for all in 1979? As soon as the topic of scientific workflows is brought up, there are always a few make fans fervently insisting that the problem of workflows is solved once and for all with GNU make , written first in the 70’s :)
Personally I haven’t been so sure. On …




