
We just wanted to share that the paper on our Go-based workflow library, SciPipe, was just published in GigaScience:
Abstract Background The complex nature of biological data has driven the development of specialized software tools. Scientific workflow management systems simplify the assembly of such tools into …

A pre-print for our Go-based workflow libarary SciPipe , is out, with the title SciPipe - A workflow library for agile development of complex and dynamic bioinformatics pipelines , co-authored by me and colleagues at pharmb.io : Martin Dahlö , Jonathan Alvarsson and Ola Spjuth . Access it here .
It has been more than …
Update (May 2019): A paper incorporating the below considerations is published:
Björn A Grüning, Samuel Lampa, Marc Vaudel, Daniel Blankenberg, “Software engineering for scientific big data analysis ” GigaScience, Volume 8, Issue 5, May 2019, giz054, https://doi.org/10.1093/gigascience/giz054 There are a …
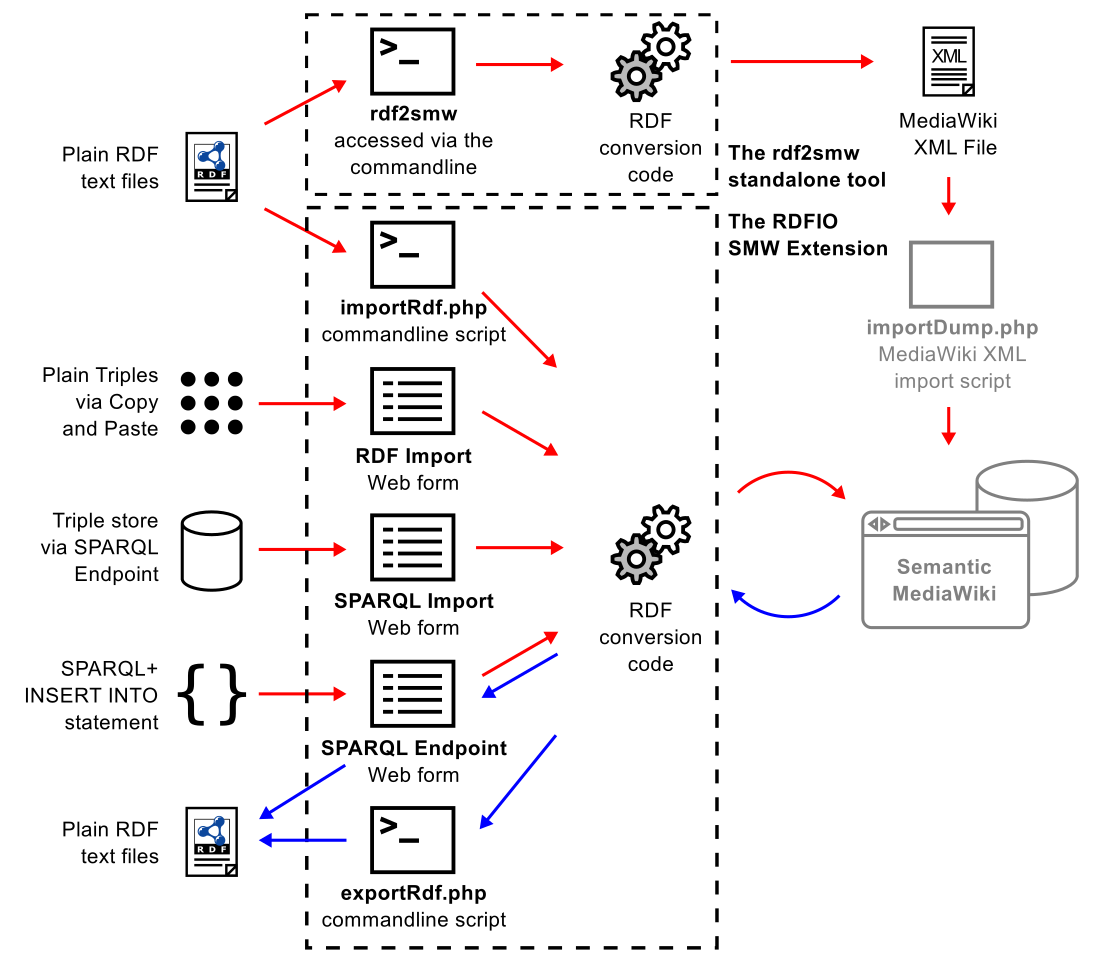
As my collaborator and M.Sc. supervisor Egon Willighagen already blogged , we just released a paper titled: “RDFIO: extending Semantic MediaWiki for interoperable biomedical data management ”, with uses cases from Egon and Pekka Kohonen , coding help from Ali King and project supervision from Denny …


