
Ad-hoc tasks in bioinformatics can contain such an immense number of operations and tasks that need to be performed to achieve a certain goal. Often these are all individually regarded as rather “standard” or “routine”. Despite this, it is quite hard to find an authoritative set of …
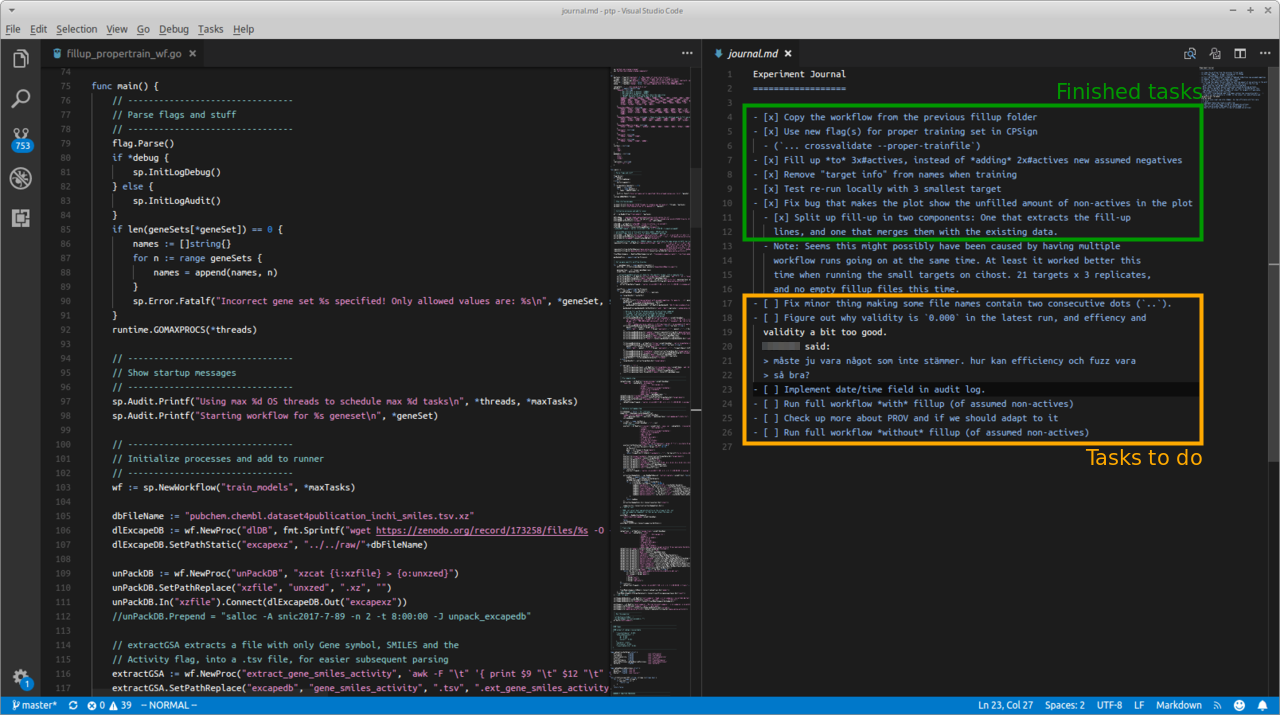
Good lab note-taking is hard Good note-taking is in my opinion as important for computational research as for wet lab research. For computational research it is much easier though to forget doing it, since you might not have a physical notebook lying on your desk staring at you, but rather might need to open a specific …
One year left to the dissertation (we hope) and now turning from mostly software development into more of data analysis and needing to read up quite a pile of books and papers on my actual topic, pharmaceutical bioinformatics. With this background, I’m feel forced to ponder ways to improving my note taking …
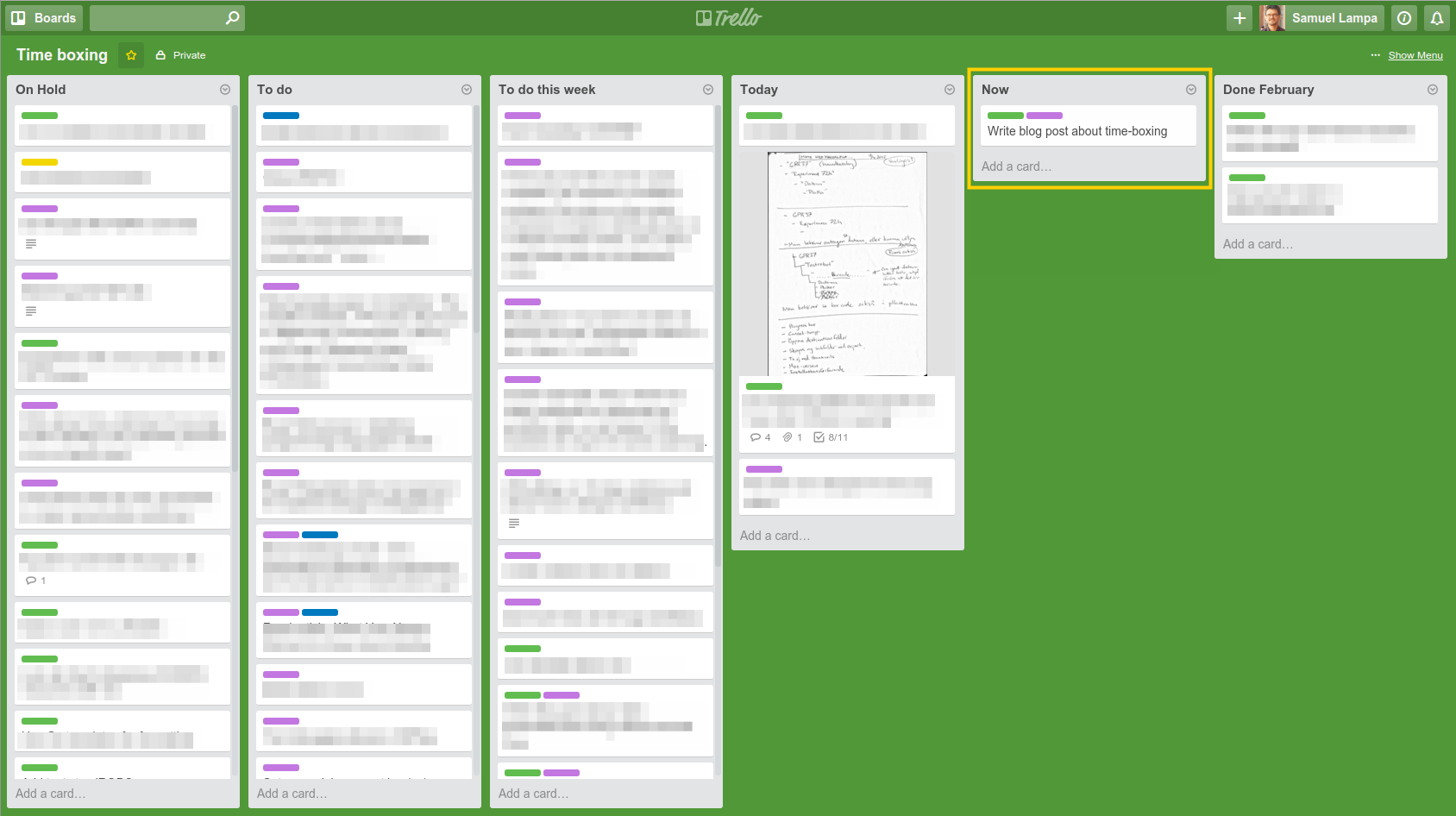
Figure: Sketchy screenshot of how my current board looks. Notice especially the “Now” stack, marked in yellow, where you are only allowed to put one single card. I used to have a very hard time getting an overview of my current work, and prioritizing and concentrating on any single task for too long. I …

I had heard a lot of people say vim is very hard to learn, and got the impression that it will take a great investment to switch to using it.
While I have came to understand that they are right in that there is a lot of things to invest in to get really great at using vim, that will really pay back, I have also found …
I have tried hard to improve my linux desktop productivity by learning to do as much as possible using keyboard shortcuts, aliases for terminal commands etc etc (I even produced an online course on linux commandline productivity ).
In this spirit, I naturally tried out a so called tiling window manager (aka tiling wm). …
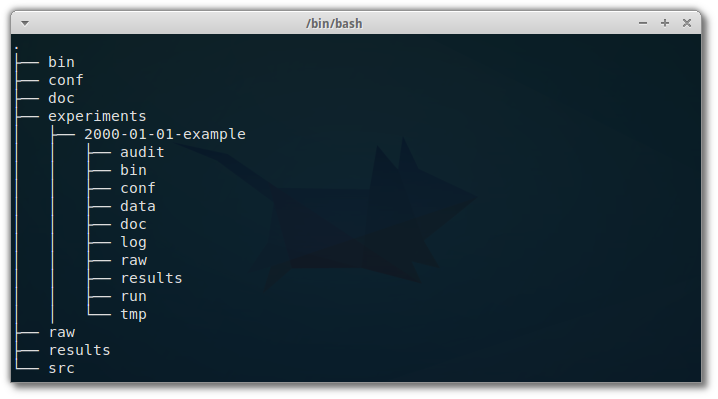
I read this excellent article with practical recommendations on how to organize a computational project, in terms of directory structure.
Directory structure matters The importance of a good directory structure seems to often be overlooked in teaching about computational biology, but can be the difference between a …
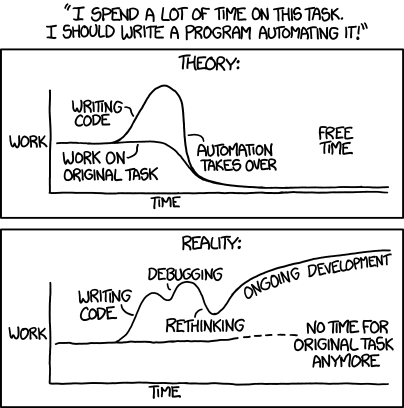
Disclaimer: Don’t take this too seriously … this is “thinking-in-progress” :)
It just struck me the other minute, how simplicity is the key theme behind two very important areas in software development, that I’ve been dabbling with quite a bit recently: Testing, and automation.
Have you …






