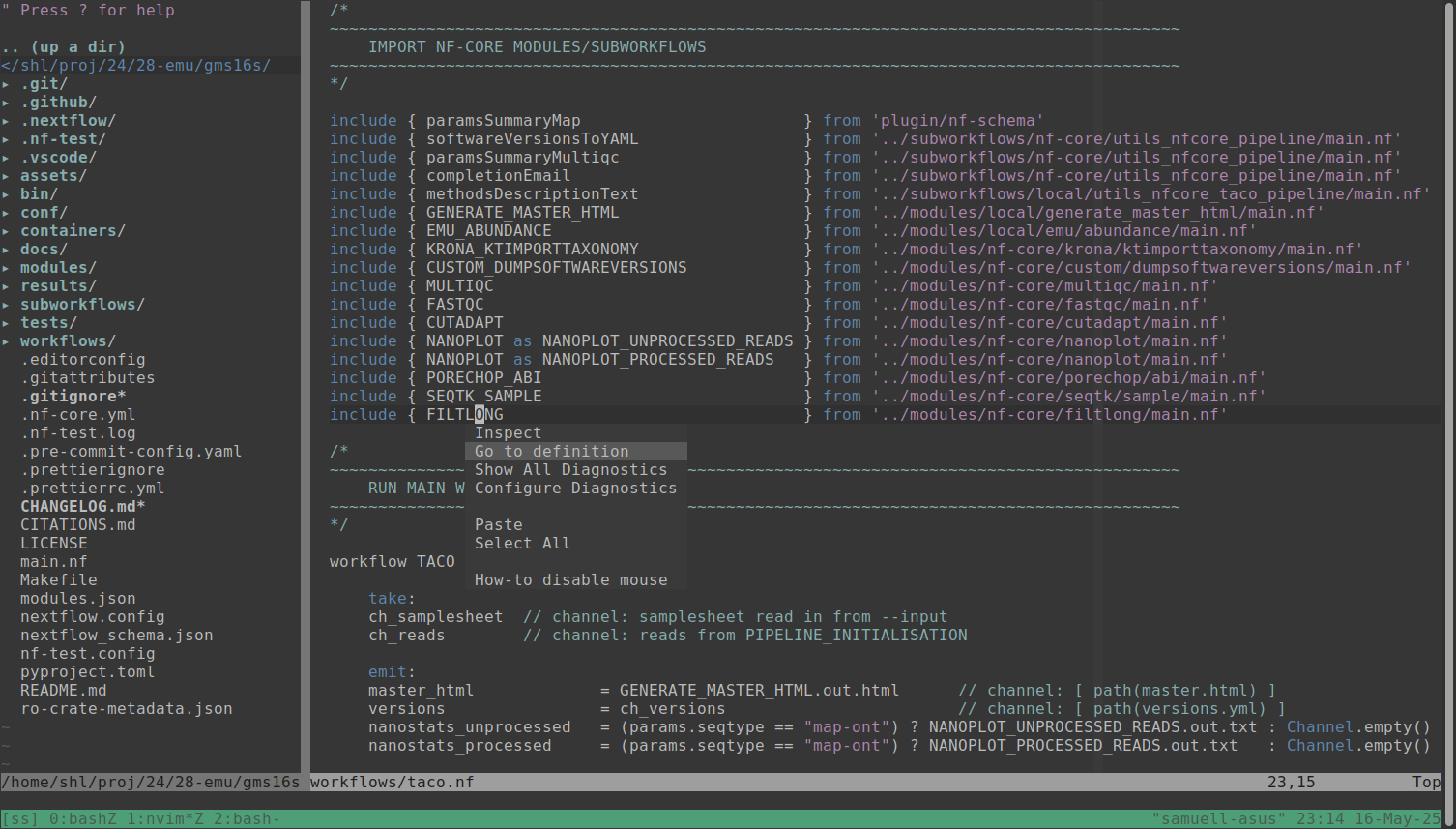
It turns out it is super easy to get Nextflow code intelligence to work in NeoVim now! This is thanks to the newly released Nextflow language server following the increasingly popular Language Server Protocol (LSP).
See this episode of the Nextflow podcast for a more in-depth discussion on this new functionality.
The …
We have been evaluating Nextflow before in my work at pharmb.io , but that was before DSL2 and the support for re-usable modules (which was one reason we needed to develop our own tools to support our challenges, as explained in the paper ). Thus, there’s definitely some stuff to get into.
Based on my years in …
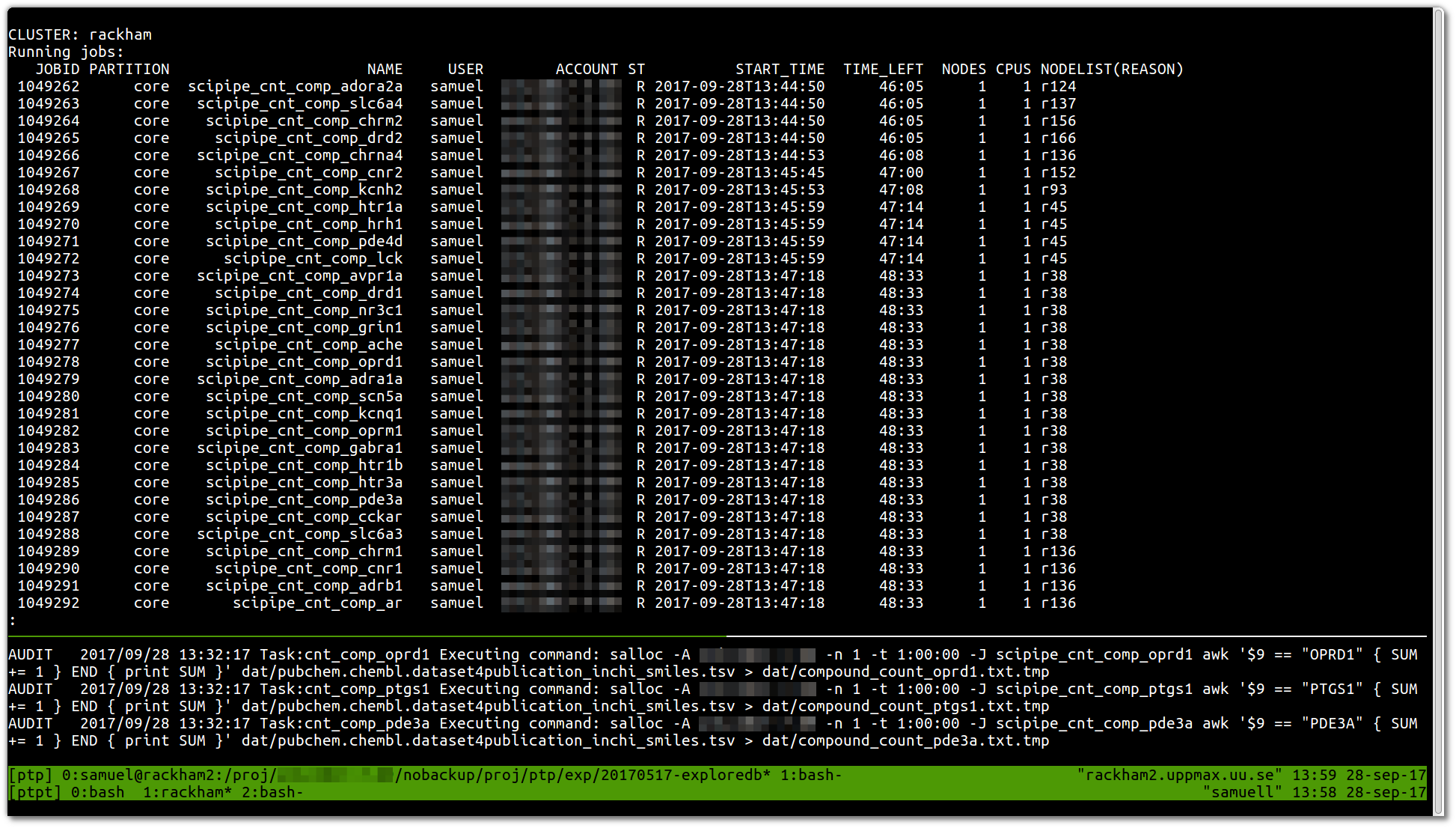
Today marked the day when we ran the very first production workflow with SciPipe , the Go -based scientific workflow tool we’ve been working on over the last couple of years. Yay! :)
This is how it looked (no fancy GUI or such yet, sorry):
The first result we got in this very very first job was a list of counts …

